PyCon was incredible!Permalink
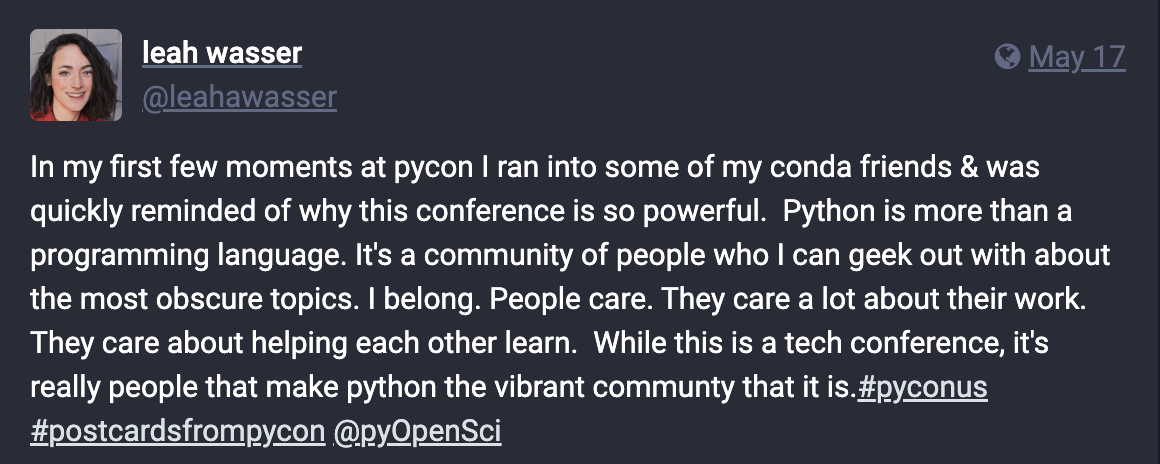
We feel so incredibly fortunate to have connected with so many of you at PyCon US! While we’re still recovering from a week of summits, talks, and sprints, we wanted to share this sentiment from our Executive Director and Founder, Leah Wasser, which she shared on her Fosstodon account:
pyOpenSci’s PyCon US Open SpacePermalink
This year, pyOpenSci’s first Open Space brought together 17 attendees from various organizations, including Anaconda, NVIDIA, CPython, and more. The discussions focused on interoperability, scientific data workflows, and Python packaging for science, emphasizing the need for consistent tools and data formats to reduce cognitive load for scientists. A key topic was the gap between exploratory scripts and fully packaged code, highlighting the need for “in between” options for sharing code and outputs. These insights will guide pyOpenSci’s efforts, with more discussions planned for the Fall Festival, scheduled for September 28-29, 2024.
pyOpenSci’s PyCon US SprintPermalink
This year we had a tremendous turnout of over 20 people from several countries for our 1-day pyConUS sprint. At last count this resulted in about 30 issues and pull requests where volunteers who were there to support scientists, helped us fix issues on our website, in our tutorials and our packaging and peer review guidebooks. So many long-standing issues and bugs were fixed thanks to this wonderful Python community.
See you at SciPyPermalink
SciPy 2024 is just around the corner, and we can’t wait to see you there! We’re pulling together our pyOpenSci Guide to SciPy, similar to the guide we did for PyCon 2024, and wanted to give you a preview of some of the tutorials and talks being given by pyOpenSci Community members!
TutorialsPermalink
Interactive data visualizations with Bokeh (in 2024)Permalink
Along with Timo Metzger and Bryan Van de Ven, pyOpenSci community member Pavithra Eswaramoorthy will be covering everything you need to know to create beautiful and powerful interactive plots from scratch using Bokeh’s latest features. Starting with a quick introduction of Bokeh’s core concepts, the team will cover creating and customizing simple static plots like line and bar charts before introducing layers of interactivity, creating specialized plotting features like geographic maps, contour plots, Mathematical Text, and discussing new additions to Bokeh like ImageStacks. By the end, you will be able to create a complete interactive dashboard using Bokeh.
3D Visualization with PyVistaPermalink
Led by Tetsuo Koyama, Alexander Kaszynski, Bill Little, and Bane Sullivan, this tutorial demonstrates PyVista’s latest capabilities and bring a wide range of users to the forefront of 3D visualization in Python, including:
- Use PyVista to create 3D visualizations from a variety of datasets in common formats.
- Overview the classes and data structures of PyVista with real-world examples.
- Be familiar of the various filters and features of PyVista.
- Know which Python libraries are used and can be used by PyVista (meshio, trimesh etc).
- We see this tutorial catering to anyone who wants to visualize data in any domain, and this ranges from basic Python users to advanced power users.
From RAGS to riches: Build an AI document inquiry web-appPermalink
Pavithra Eswaramoorthy, a Developer Advocate at Quansight, is teaming up with Dharhas Pothina and Andrew Huang to cover how to use RAG to build document-inquiry chat systems using different commercial and locally running LLMs. The topics we’ll cover include:
- Introduction to RAG, how it works and interacts with LLMs, and Ragna - a framework for RAG orchestration
- Creating and optimizing a basic chat function that uses popular LLMs (like GPT) answers questions about your documents, using a Python API in Jupyter Notebooks
- Running a local LLM on GPUs on the provided platform, and comparing its performance to commercial LLMs
- Introduction to Panel, by creating a basic chat UI for Ragna using Panel’s ChatBox widget
- Building and deploying a Panel-based web-app, which extends the basic chat UI and includes more application components
By the end of this tutorial, you will have an understanding of the fundamental components that form a RAG model, and practical knowledge of open source tools that can help you or your organization explore and build on your own applications. This tutorial is designed to enable enthusiasts in our community to explore an interesting topic using some beginner-friendly Python libraries.
Create Your First Python Package: Make Your Python Code Easier to Share and UsePermalink
Led by pyOpenSci’s Executive Director and Founder, Leah Wasser, in this tutorial session you’ll learn how to:
- Create code that can be installed into different environments
- Use Hatch as a workflow tool, making setup and installation of your code easier
- Use Hatch to publish your package to (test) PyPI
Creating code that can be shared and reused is the gold-standard of open science. But tools and skills to share your code can be tricky to learn. In this hands-on tutorial, you’ll learn how to turn your code into an installable Python module ( a file containing Python code that defines functions, classes, and variables), that can be shared with others. We will provide pre-built code and module examples for each step of the tutorial to make it more beginner-friendly. However, to get the most out of this tutorial, you should be familiar with writing Python code, writing and using functions and you should understand the concept of Python environments. You should also have Python and Hatch installed on your computer prior to attending. If you are newer to environments and don’t have a preferred tool, then we recommend that you use conda. However, any environment tool that you prefer works well.
Interested in helping out with our Packaging workshop? Reach out to us at media@pyopensci.org. We’re looking for volunteers to assist learners throughout the workshop.
Data of an Unusual Size (2024 edition): A practical guide to analysis and interactive visualization of massive datasetsPermalink
Pavithra Eswaramoorthy, and Dharhas Pothina, both from Quansight, will help you learn the fundamentals of analyzing massive datasets with real-world examples on actual powerful machines on a public cloud provided by the presenters – starting from how the data is stored and read, to how it is processed and visualized.
TalksPermalink
The power of community in solving scientific Python’s most challenging problemsPermalink
This talk from our Executive Director and Founder, Leah Wasser, shares pyOpenSci’s knowledge in building a constructive open peer review process that supports maintainers, and also resources that make Python packaging more accessible and easier-to-navigate. Our insights are based on five years of experience working with over 200 community members, who have evaluated 58 packages developed by over 56 package maintainers.
Leah’s talk is about writing the pyOpenSci Python Packaging Guide via community!
Great Tables for EveryonePermalink
This talk from Rich Iannone, whose package Great Tables is currently under review with pyOpenSci, will provide a demonstration of Great Tables, showing how data can come alive in when in tabular form. We’ll use data from the fields of meteorology, chemistry, and the atmospheric sciences within our table-making examples. We’ll learn how the different components of a table (e.g., the column labels, the header, etc.) can be manipulated to best convey the data. We believe tables are worthy of being deemed data visualizations and we hope that Great Tables can become an indispensable tool in your data analysis workflow.
From Code to Clarity: Using Quarto for Python DocumentationPermalink
Isabel Zimmerman, a member of our triage team and an Emeritus Editor in Chief, will be speaking from her experiences as an open source developer who works with multilingual tools, and how Quarto + quartodoc helped to fill those gaps. This talk is intended for anyone who might have to communicate with stakeholders through code using a web-based format. She’ll be focusing on Python package documentation websites, but the stories and examples expose the difficulties in many types of technical communication. The intent is to have a lighthearted talk filled with examples of how to make technical communication and publishing easier and more accessible.
Vector space embeddings and data maps for cyber defensePermalink
pyOpenSci community member Benoit Hamelin will cover how the vast amounts of information of interest to cyber defense organizations comes in the form of unstructured data; from host-based telemetry and malware binaries, to phishing emails and network packet sequences. All of this data is extremely challenging to analyze. In recent years there have been huge advances in the methodology for converting unstructured media into vectors. However, leveraging such techniques for cyber defense data remains a challenge.
Imposing structure on unstructured data allows us to leverage powerful data science and machine learning tools. Structure can be imposed in multiple ways, but vector space representations, with a meaningful distance measure, have proven to be one of the most fruitful.
In this talk, he will demonstrate a number of techniques for embedding cyber defense data into vector spaces, as well as discuss how to leverage manifold learning techniques, clustering, and interactive data visualization to broaden our understanding of the data and enrich it with expert feedback.
SprintsPermalink
We’ll also be holding a pyOpenSci sprint at SciPy 2024! Be sure to follow us on LinkedIn and Fosstodon for more details!
Save the Date: pyOpenSci’s Open Source Fall FestivalPermalink

We love a good get-together, and our Open Source Fall Festival is no exception! Held entirely online using Gather, our goals is to bring together members of the Python community that create open source tools with the members of the Python community who use the tools. In building this festival, our focus is on a grassroots, community-led event with some big unconference vibes. So mark your calendars, and save the date for September 28–29th, 2024!
Python packages for everyone!Permalink
pyOpenSci’s Open Peer Review process is experiencing an unprecedented number of Python package submissions! We’re excited to see so many wonderful packages that are helping scientists help make the world a better place being submitted for review. Our open peer review process facilitates scientists getting credit and recognition for the work they’ve invested in developing scientific Python tools while also supporting them in building better software. The peer review process also supports scientists in finding vetted and maintained software, which drives their open science workflows.
You can submit your package for review (or pre-submission inquiry to determine if it’s in scope) by submitting an issue to our software-submission repository. And because all of our reviews are open, you can see our review process in action.
We’re also accepting applications for package reviewers. We welcome reviewers from a diversity of background and with varying levels of Python technical expertise.
Some of the basic things that we look for in a review include:
- Familiarity with using the Python programming language.
- Ability to evaluate a Python package for usability and documentation quality.
- Ability to provide a technical review of Python package structure and code quality / approach to solving the problems that the package seeks to address.
We like to have a mix of technical and usability focus in our reviews so it’s ok if you don’t have all of the above skills! Not sure if you’re ready to review on your own? Check out the pyOpenSci mentorship program!
Get involved with pyOpenSci: call for editorsPermalink

If you’re looking to give back to the broader Python community, consider joining our Editorial Board! The pyOpenSci Editorial Board is comprised of a diverse group of volunteer editors, each one responsible for the following tasks:
- Finding reviewers from diverse backgrounds who have a mixture of scientific domain and Python experience.
- Overseeing the entire review process for a package ensuring it runs in a timely and efficient manner.
- Supporting the submitting authors and reviewers in answering questions related to the review.
- Determining whether that package should be accepted into the pyOpenSci ecosystem once the review has wrapped up.
While we have a current need for editors with expertise in climatology and/or energy, we’re also recruiting editors from all scientific disciplines. If you feel you’d be a good fit for the pyOpenSci Editorial Board, please fill out this form.
If you’re interested in joining our Editorial Board, but have questions or would like to chat with someone from pyOpenSci about volunteering to be an editor, please email us at media@pyopensci.org.
Upcoming Python events for scientistsPermalink
SciPyPermalink
With keynotes from Julia Silge, Dr. Elizabeth (Libby) Barnes, and Kyle Cranmer, SciPy 2024 is going to be yet another conference you don’t want to miss! Held at the Tacoma Convention Center, July 8-14, 2024, tickets are still available!
PyData EindhovenPermalink
With 14 captivating talks lined up, PyData Eindhoven offers a diverse range of topics to expand your knowledge and inspire innovative thinking. From cutting-edge data analysis techniques to machine learning applications, our expert speakers will cover it all. Co-hosted with Day 2 of JuliaCon, grab your tickets and join your peers in the Netherlands this July 11th!
BioC 2024Permalink
Taking place Wednesday July 24 - Friday July 26 at the Van Andel Institute in Grand Rapids, MI,the BioC 2024: Where Software and Biology Connect hybrid conference still has tickets available. And what’s more, all proceeds go to helping underrepresented minorities and individuals from low to middle-income countries, including students, fellows, and early career investigators.
PyData VermontPermalink
PyData Vermont 2024 is a two day in-person event for the international community of data scientists, data engineers, and developers of data analysis tools to share ideas and learn from each other. The event will be in-person at the Main Street Landing Performing Arts Center, July 29 to July 30, 2024. Register today!
Tell us about your event!Permalink
Have an upcoming scientific Python event that’s open to the public? Email us at media@pyopensci.org with the details to have your event listed.
Connect with usPermalink
Stay up-to-date with all things pyOpenSci by following us on LinkedIn and Fosstodon, and you can connect with the broader pyOpenSci community on our Discourse forum. And if you’re interested in our weekly newsletter where we share news, blog posts, and monthly updates, subscribe on LinkedIn. We also send out a monthly recap newsletter to our mailing list!

Leave a comment